A large-scale Atlas of Human Neural Organoid Cells to map the fetal and postnatal brain using a common coordinate framework
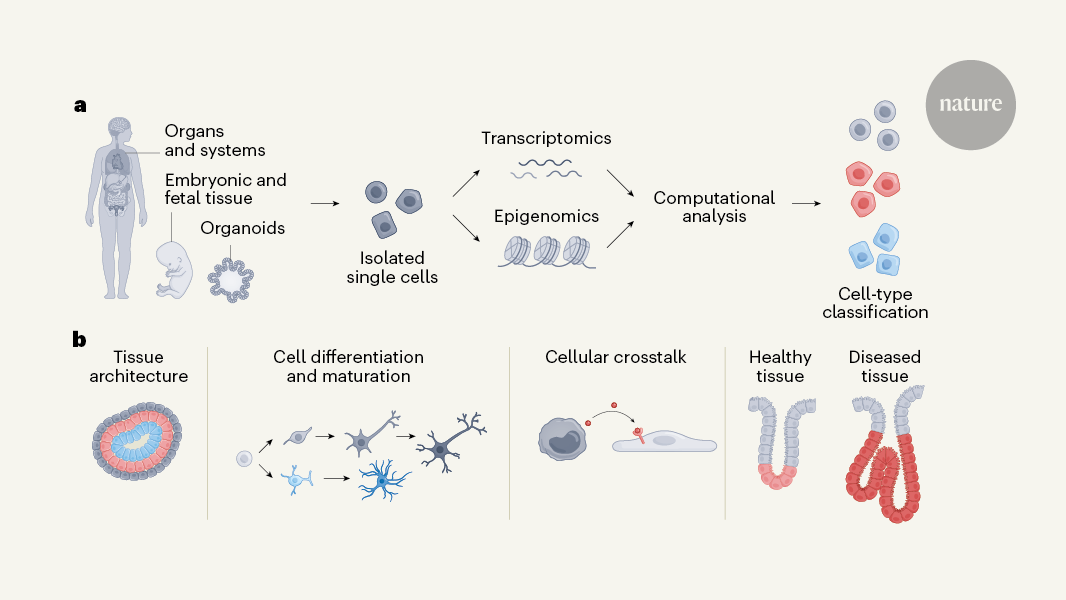
The atlases rely heavily on the characterization of many individual cells and their RNA transcripts (their transcriptomes), owing to both technological developments and a key biological insight. First, over the past ten years, technology has scaled up to allow scientists to capture the transcriptomes of many thousands of cells in a single experiment. Over the past 30 years of research, the transcriptome has been able to capture cell types and cell states.
The gastrointestinal tract atlas by Oliver et al.2 covers the entire pharoah, stomach, and colon. A large-scale atlas of more than one million cells is being created using an innovative computational approach, which integrates previous data sets. The authors have a lot of data sets from individuals with inflammatory diseases, including coeliac disease and Crohn’s disease. Intestinal inflammation can cause cells to undergo metaplasia, a shift from one cell type to another. The authors inferred the origin of the metaplastic cells by looking at the data sets and comparing them with stem cells. The benefit of the atlas’s completeness is that it allows a comparison of disease states in one organ and normal states of cells in another.
Organoids are a powerful model for functional analysis because they are so easy to study. The human neural organoid cell atlas was built on 1.7 million cells and was underpinned by 36 single-cell RNA-sequencing data sets. The question of how faithfully organoids capture aspects of the developing brain is already being answered by the atlas. The organoids were in culture for three months, and they looked like the brain of a fetal during the first three weeks of the fetus’s life. But the authors found an intriguing limit to this correspondence. The fetal brain during the last trimester of pregnancy isn’t captured in organoids and this leaves an open question as to what signals are missing from these models.
Yayon et al.4 created a map of the thymus, a lymphoid organ that produces immune cells, in its early fetal development and early postnatal stages. Using the spatial dimension, the authors conceived of a ‘common coordinate framework’ to mathematically map the tissue. This model of the axis between the outer part of the thymus and its centre (the cortico-medullary axis) allows for a deeper understanding of tissue organization and comparison of the organ both in and between individuals. It will be interesting to study how the Atlas extends to other stages of life.
Cells are continuously differentiated over space and time in embryo development, which leads to the development of tissues, organs and the establishment of body functions. Scientists’ understanding of the cellular mechanisms that underlie such events remains limited due to the immense complexity.
The development of the skull and joints that make up the limbs in the first few weeks after conception was studied. They mapped transcriptomic and epigenomic profiles of single cells and identified key gene-regulatory networks that directed the commitment of cells to chondrogenic and osteogenic regions. The authors inferred probable lineage relationships along differentiation pathways and propose how cellular crosstalk might guide the formation of bone, identifying a potential key role for interactions with the vascular system. An elegant approach has been used by the authors to combine data from genome-wide association studies with their single-cell analysis to find cell states that are potentially related to osteoarthritis, a disease affecting the joints.
Gopee and colleagues present a comprehensive atlas of skin development after 7 weeks of conception. Using a combination of transcriptomic technologies the authors mapped changes in cell states and showed how these organize to form different types of structures. Their research shows the unexpected role of immune cells in coordinating the development of blood vessels and is also related to the formation of blood vessels. This was further validated through an innovative organoid system that recapitulates key aspects of skin development.
sc Tab is a deep-learning model that can be tailored to annotating cell types in different tissues. Recognizing the limitations of conventional machine learning in handling large, diverse data sets, the developers of scTab introduce a data-augmentation approach for single-cell sequencing data to increase the size of the training data set, enabling generalizations to be made across tissues. If trained on extensive data sets, complex equations beat simpler equations in cell-type classification. The potential of scTab lies in promoting standardized cell labels and encouraging a research-community-wide consensus in cell-type nomenclature.
MultiDGD is a way to integrate multimodal data such as genes expression and accessibility of chromatin using a deep variable model. This type of machine learning uses hidden variables to learn complex patterns — in this case, multiDGD learns optimal hidden-variable representations that are shared across multiple data modalities (transcriptomic and epigenomic), without the need to pre-define important features. It’s suitable for multi-omics studies in which data were gathered from different sources if the information about potentially confounding variables is included in multiDGD. The models clustering of shared representations improves the alignment of data, which is important to understand the genes and regulatory regions in the genome.
PopV and sc Tab lead efforts in standardized annotations and consensus-building, while multiDGD opens up possibilities for data integration across complex multimodal data sets.
This shows the importance of innovation and the rapid pace of the field. To meet this growth, future research is likely to emphasize adaptable and interoperable solutions. These methods contribute valuable foundations for future advancements, paving the way for even more adaptable and scalable models for single-cell, multi-omics data.
Towards a sustainable global human cell Atlas: Challenges and Opportunities for future research in the lung cell atlas, and it’s remarkable
The studies from the global project are a major achievement, almost a decade after it was launched. Funders should sign up for the long haul.
Findings from researchers working on the lung cell atlas, for example, highlight the differences between the lungs of a sample of people in Malawi who died from COVID-19 and those who died from other lung diseases2. Scientists have been analyzing the development of human skin and joints during the second and third years of life.
The HCA was possible because of the earlierprojects, including the Human Genome Project and the National Institute for Diabetes and Tuberculosis BRAIN Initiative. HCA teams work hard to represent human diversity in their data. The scientists from Africa, Asia, Latin America and the Middle East are all part of the consortium. Researchers were invited to join, but also to help coordinate and lead HCA projects according to priorities relevant to local populations. The initiative now involves more than 3,000 scientists across some 1,700 institutions, recording and studying data from people in around 100 countries.
Most research projects — including those involving large-scale consortiums — have a limited lifespan. Ten years is considered generous. A handful of projects might last a few years longer. The tools and technologies without which important discoveries and inventions would be impossible are usually the projects which get the permanent funding. That is what the HCA needs to be compared to.
What is the Majority of Disease-Associated Variants in a Human Genome? And What Are They Really Trying to Decipher?
“While genetic studies have mapped more than 100,000 disease-associated variants in the human genome, we do not know in which cells the majority of these variants are active,” HCA researchers write6. Without this knowledge, they add, “we cannot fully understand biology, study more powerful models of disease, deploy better diagnostics, and develop more effective therapies”.