In search of the sex of the remains from the Chichén Itzá shrine: a genetic study of close relatives and twins
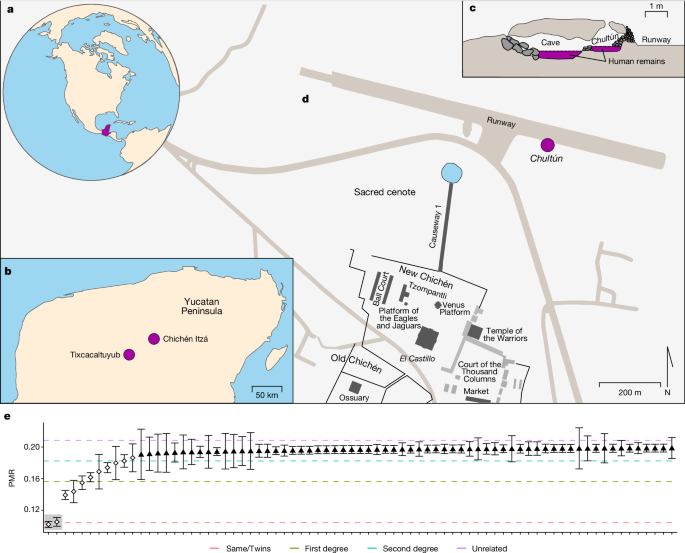
“This was very, very astonishing,” says study co-author Oana Del Castillo-Chávez, a biological anthropologist at the Yucatán National Institute of Anthropology and History in Mérida, Mexico. Remains from the Sacred Cenote included those of boys and girls, and there is no evidence from Chichén Itzá or other ancient Maya cities of close relatives being sacrificed, she adds.
The children Del Castillo-Chávez and her colleagues analysed were found in the 1960s in an underground chamber called a chultún and an adjacent cave, near the Sacred Cenote. The remains were found as part of a shrine and there were no signs of violence.
In the hope of identifying the sex of the remains and to glean other genetic insights, Del Castillo-Chávez teamed up with immunogeneticist Rodrigo Barquera and palaeogeneticist Johannes Krause at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, and their colleagues. The ancient genome data was obtained from 64 of the 106 people who were buried at the chultn.
The children were sacrificed between the seventh and mid-twelfth century ad, radiocarbon dating suggested. The genome data showed that 25% had a sibling or cousin, and that two pairs of identical twins were among the victims. The presence of twins and close relatives could be linked to rituals that involved twin figures from Mayan mythology, the researchers suggest.
Barquera says it was probably a part of preparing them for the sacrifice. “Death and sacrifice for them means something completely different to what it means to us. For them, it was a big honour to be part of this.”
A comparative analysis of the relationship between the Salmonella enterica sp. paratyphi allgene and the cocoliztli epidemic
There is a link between an HLA allgene that has become more common and protection against severe Salmonella infections. A previous study by Krause’s team has linked the bacterium Salmonella enterica sp. Paratyphi to a sixteenth-century disease outbreak called the cocoliztli epidemic4, which killed millions of people in Mexico and beyond.
To assess the genetic relationships and patterns of admixtures suggested by the PCA and ADMIXTURE analysis, we carried out F-statistics analyses using the Xerxes CLI software (v.0.3.0.0) from the Poseidon (v.2.5.0) framework162. When the divergence from an African outgroup occurs, we calculated a number for the amount of shared genetic drift from the target populations to the outgroup, where X was either YCH or TIX. We then used a subset of Native American populations (those with the top F3 scores from our previous test) to perform F4 tests of the form f4(Mbuti, YCH; test, TIX) to investigate if the YCH and TIX groups are more closely related to each other or shared an excess of alleles with any population in position test. A negative value means that either YCH and test or TIX and Mbuti share more all genes than anticipated under the null hypothesis. On the other hand, a positive value suggests that YCH and TIX share an excess of alleles between themselves. To test whether the newly sequenced individuals were genetically similar enough to be grouped into one group, we used qpWave50 v.420, a modelling approach which evaluates the likelihood of two individuals to derive from the same ancestry, relative to a given set of references including worldwide populations. A value below 0.05 indicates a low likelihood, from which we conclude that one of the individuals has a different ancestry profile. The individuals are all the same in ancestry, and that’s consistent with most tests. The person is the director of Onge. DG, Papuan. Russia_MA1_HG.SG, USA_Ancient_Beringian are the name of the directors. SG, USA_Anzick. SG, Mixe.DG, Mexico_Zapotec. DG, Belize_MayahakCabPek_9300BP, Karitiana. The director of Piapoco. DG, Peru_LaGalgada_4100BP, Pima, Cabecar. There are few exceptions, in which an individual shows low likelihood of sharing ancestry with several other individuals. Some people produced higher rates of pairs with a low likelihood. To investigate which ancestries differentiate the respective individual from most individuals, we calculated the differential affinities using an F4 statistics of the form f4(Mbuti. In which X would be the respective individual and Y present-day groups and published ancient groups and individuals. If the test group in Y has a higher affinity to the tested individual than the group in X, we would assume a positive test score. There is no modern group or individual that could explain the behavior in the qpWave analysis. We tested if present-day speakers of Mayan were consistent with their ancestors from the ancient people of Chichén Itz. Onge is the director of Onge. DG, Papuan. DG, Han.DG, Russia_MA1_HG.SG, USA_Ancient_Beringian. SG, USA_Anzick. SG, Mixe.DG, Mexico_Zapotec. DG, Belize_MayahakCabPek_9300BP and Karitiana. DG. The ancient and present-day groups have the same ancestry, as shown by the resulting P values. The same test was not successful for other present-day populations such as Gen-Pano-Carib or Chibchan-Paezan speakers as well as for the seemingly unadmixed individuals from Tixcacaltuyub, possibly attributable to European admixture not detected in the model-based clustering analyses (Fig. 2 and Extended Data Fig. 1). When modelling their ancestry composition relative to the populations Mbuti. The director of Onge. The man is from Papuan. Russia_MA1_H.SG, USA_Ancient_ Beringian are the directors of the company. SG, USA_Anzick. SG, Mixe.DG, Mexico_Zapotec. Maya HakCabPek, the director, is based in the country of Belize. DG as a mixture of Yoruba. DG, Spanish. The working model suggests a composition of mostly Mexican, and mainly Spanish, people.
We mapped sequence data from the immunity-captured libraries against a custom panel containing 1,225 allelics with a ‘common’ or ‘intermediate’ CIWD 3.0 designation. The number of matches was set to zero. In this way we obtained HLA class I and class II alleles for the genes HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1. Of the 882 raw allele calls, 72 (8.1%) Secondary allele predictions were overruled in favour of anomalies with a read cross-mapped to several genes. The frequencies and properties reported were used to assign haplotypes. When possible. To compare the HLA frequencies between precontact Chichén Itzá and postcontact Tixcacaltuyub, we performed Fisher exact tests154 and adjusted the P values for several comparisons according to the Benjamini–Hochberg correction155 which control the false discovery rate (FDR). The FDR approach controls for a very low proportion of false positives, instead of ensuring that there are no false positives, which results in increased statistical power. It has been argued74 that certain types of non-overlapping association between HLA loci may be a signature of pathogen selection. The (f_rmadj ) metric was calculated by using the YCH and TIX haplotypes for which we had data for every HLA locus. To provide a point of comparison for the ({f}_{{\rm{adj}}}^{ }) score of each locus pair for both YCH and TIX, we generated 5,000 random permutations of the order of the total alleles at one of the loci in each pair and recalculated ({f}{{\rm{adj}}}^{ }) for each set of randomized data. We generated distributions of possible ({f}_{{\rm{adj}}}^{ }) scores for each pair of loci in the dataset that would be obtained if the alleles at those loci were associated entirely at random. We then calculated the difference between the ({f}{{\rm{adj}}}^{ }) value calculated from each dataset for each pair of loci and the mean of the ({f}_{{\rm{adj}}}^{ }) scores calculated from the randomized data for that pair of loci, then divided that difference by the standard deviation of ({f}_{{\rm{adj}}}^{* }) calculated from the randomized data. We were able to rank the pairs based on how much non-overlap they showed relative to random associations. To further assess the role of HLA class II alleles in resistance to S. enterica infection, we ran in silico binding predictions for the HLA-DRB1/3/4/5 alleles and -DQA1/DQB1 allele pairs found in YCH individuals and peptides derived from 18 Salmonella spp. proteins that have been previously reported to be highly immunogenic in humans (Supplementary Methods: ‘In silico binding prediction assays’) using NetMHCIIpan-4.0 (ref. 82) as implemented in the IEDB Analysis Resource virtual machine image157,158. On the basis of adjusted rank values, binder’s were classified into strong and weak depending on their rank.
The haplogroups were mapped to the revised Cambridge reference sequence. For the resulting sequences, we used HaploGrep2 (ref. 148) and HAPLOFIND149 to assign and confirm the corresponding mtDNA haplogroups (Supplementary Fig. 6). For the Y-haplogroup assignments, we created pileups of reads for each individual which mapped to Y chromosome (Y-Chr) SNPs as listed on the ISOGG Y-DNA Haplogroup Tree (v.15.73; https://isogg.org/tree/). We then manually assigned Y-Chr haplogroups for each individual based on the most downstream SNP retrieved after evaluating the presence of upstream mutations along the Y-Chr haplogroup phylogeny (Supplementary Fig. 6). There are two tables that contain full haplogroups and Y-Chr genes.
A panel of over 1 million SNPs were enriched from the total DNA in the libraries, using an in-solution capture approach. A portion of captured library pools were then mapped on the Illumina Hiseq 4000 platform for a total of 20 million reads.
We built non-UDG-treated libraries using 15 μl of each YCH extract to assess the authenticity of the extracted DNA after obtaining the characteristic damage plots associated with aDNA129. The libraries were built using 20 l of Y CH extract and the Illumina-specific adaptors after modifying the library preparation protocol. We built non-UDG-treated libraries for TIX extracts using 20 μl of each modern DNA extract. Both YCH and TIX libraries were quantified using quantitative PCR (qPCR) with the IS7 and IS8 primers in a quantification assay using a DyNAmo SYBR Green qPCR Kit (Thermo Fisher Scientific) on the LightCycler 96 platform (Roche Diagnostics).
For YCH, the bone powder was decalcified and proteins were digested by an overnight incubation (more than 16 h) at 37 °C in a buffer containing EDTA and Proteinase K30. The DNA was purified from the supernatant by a silica column-based method using a silica column for high volumes assay (High Pure Viral Nucleic Acid Large Volume Kit, Roche Molecular Systems). The genetic material was eluted in 100 l of TET. One mM of tris. EDTA and 0.05% Tween) and frozen at −20 °C until library preparation128. For the contemporary participants, 5 ml of peripheral blood were collected in BD Vacutainer blood collection tubes containing K2-EDTA (Becton, Dickinson and Company) and the DNA was extracted using the Quick-DNA Miniprep Plus kit (Zymo Research Corporation) following the developers’ instructions. Because our protocols are optimized for short-length aDNA and to avoid potential bias through laboratory methods, we sheared the DNA extracted from modern individuals using ultrasonic DNA shearing to an average length comparable to that of aDNA. Therefore, 50 μl of a 50 ng μl−1 dilution of each of the modern DNA samples were sheared to an average fragment length of 150 base pairs using a Covaris M220 Focused ultrasonicator (Covaris).
Direct insight into dietary trends among past populations enables the investigation of connections between diet and social status, cultural customs linked to food, environmental impacts on subsistence and perhaps even individual mobility122,123. Measurements of bone collagen δ13C and δ15N disproportionately reflect the protein component of an individual’s diet during the period of tissue formation and, to a lesser extent, the lipid and carbohydrate sources124. We analysed the bone tissue from temporal and petarial bones to reconstruct the diet of the subadult individuals from Chichén Itz. Collagen was extracted using a standard procedure125 (Supplementary Methods). The atomic C:N ratio along with the collagen yields were used to determine the quality of collagen preservation. Collagen yields of more than 1% in weight are considered acceptable for carbon and nitrogen values126, whereas the C:N ratio should range from 2.9 to 3.6 for archaeological samples127 (Supplementary Table 1).
The dog bone of 26 Y CH samples were analysed and pre treated using a standardized procedure. Collagen was extracted from the bone samples (approximately 1 g, using a modified version of the Longin method118), purified by ultrafiltration (fraction greater than 30 kD) and freeze-dried. The collagen was then combusted to CO2 in an elemental analyser. The CO2 was then converted catalytically to graphite and analysed using a MICADAS-type AMS system (Ionplus AG). The blanks, control standards and the calibration standard were all measured at the same time. A normalized 14C-age is found by using the dataset IntCal20 and the software Oxcal.
Source: Ancient genomes reveal insights into ritual life at Chichén Itzá
Collaboration and community engagement activities in the community of Tixcacaltuyub (Guadalajara, Guatemala)
In 2004, the community of Tixcacaltuyub self-identifies as a Mayan community and has been in a long term relationship with the chemistry and nursing Faculties of the University of Guadalajara. Collaboration with the community has been part of the work done by J.C.L.R., M.E.M. and J.C.T.R. As a result of these interventions, healthcare barriers and opportunities have been identified and work is now underway with the active participation of the community to implement a co-responsible model of healthcare. In April 2023, J.C.L.R. and R.B. visited Tixcacaltuyub to hold meetings with participants and elementary, high school and UADY students to return the results of the genetic findings and to collect community feedback, including how they reconcile these results with their own views. Feedback from these engagements has been incorporated into the final manuscript (Supplementary Methods: ‘Community engagement activities’). The main manuscript was translated into Spanish, as part of our strategies to make our results available to more people.